Research Summary
We are a bioinformatics lab developing computation technology to model how cell identity is established during differentiation. We develop bioinformatics methods to understand genetic, epigenetic, and molecular signaling mechanisms in cell identity regulation. Different cell types in a healthy body share the same genetic sequence. Difference in identity of these cell types is determined by epigenetic regulation of gene expression, which represents an consequence of the interaction between the genetic factors and signaling molecules in the cell. Genetic mutations and abnormal signals can lead to epigenetic alteration and cause cell identity dysregulation in diseases.

Major research approach in the lab:
-
Develop new bioinformatics techniques to analyze high throughput genomic, epigenomic, transcriptomic, and metabolomic data.
-
Single cell, spatial genomics, Hi-C, ChIP-seq, ATAC-seq, RNA-seq and other additional techniques to study epigenetic regulation of transcription by 3D chromatin conformation, chromatin modifications, and chromatin-binding proteins.
-
High throughput profiling of RNA methylation, RNA binding proteins, and ribosomes to study post transcriptional regulation of gene expression.
-
Collaborate with wet-bench labs for molecular, cellular, animal, and clinical investigations to understand mechanisms in diseases.
Research in the lab is centered around one fundamental insight into basic biology: the regulation of cell identity genes exhibits a higher degree of complexity compared to many other genes, such as housekeeping genes. Our interest in this phenomenon starts from computational modeling showing that the H3K4me3 modification breadth on chromatin follows a bimodal distribution (Nature Genetics, 2015). Specifically, cell identity genes within a cell tend to exhibit a broad H3K4me3 pattern, spanning both the promoter and gene body regions. For instance, the CD4 locus displays a broad H3K4me3 pattern in CD4+ T cells. Conversely, the majority (~95%) of expressed genes, including housekeeping genes, exhibit a sharp H3K4me3 pattern solely at the promoter. This observation echoes several findings from other laboratories, such as the Brunet lab’s work on H3K4me3 breadth (Cell, 2014) and the Young Lab’s research on super-enhancers (Cell, 2013). Furthermore, our research has revealed that cell identity genes tend to be regulated by a subset of super-enhancers marked by CTCF binding (published in GPB, 2021) and exhibit low RNA stability due to co-transcriptional RNA m6A modification (Nucleic Acids Research, 2023). These findings collectively demonstrate that cell identity genes experience a more rapid turnover in expression, encompassing both frequent transcription and mRNA decay, compared to other genes expressed at a similar level. Motivated by these findings, we developed CEFCIG, an artificial intelligence technology designed to identify cell identity genes based on the intricate nature of their expression regulation (Nature Communications, 2020). As a proof of concept, our application of this novel technology to endothelial cells has unveiled MECOM as a regulator of endothelial cell lineage (Nature Communications, 2020; Nature Communications, 2023). This research direction holds significant implications for a wide array of research communities, addressing critical challenges of cell identity conversion technologies and overcome a crucial obstacles hindering the advancement of cell therapy.
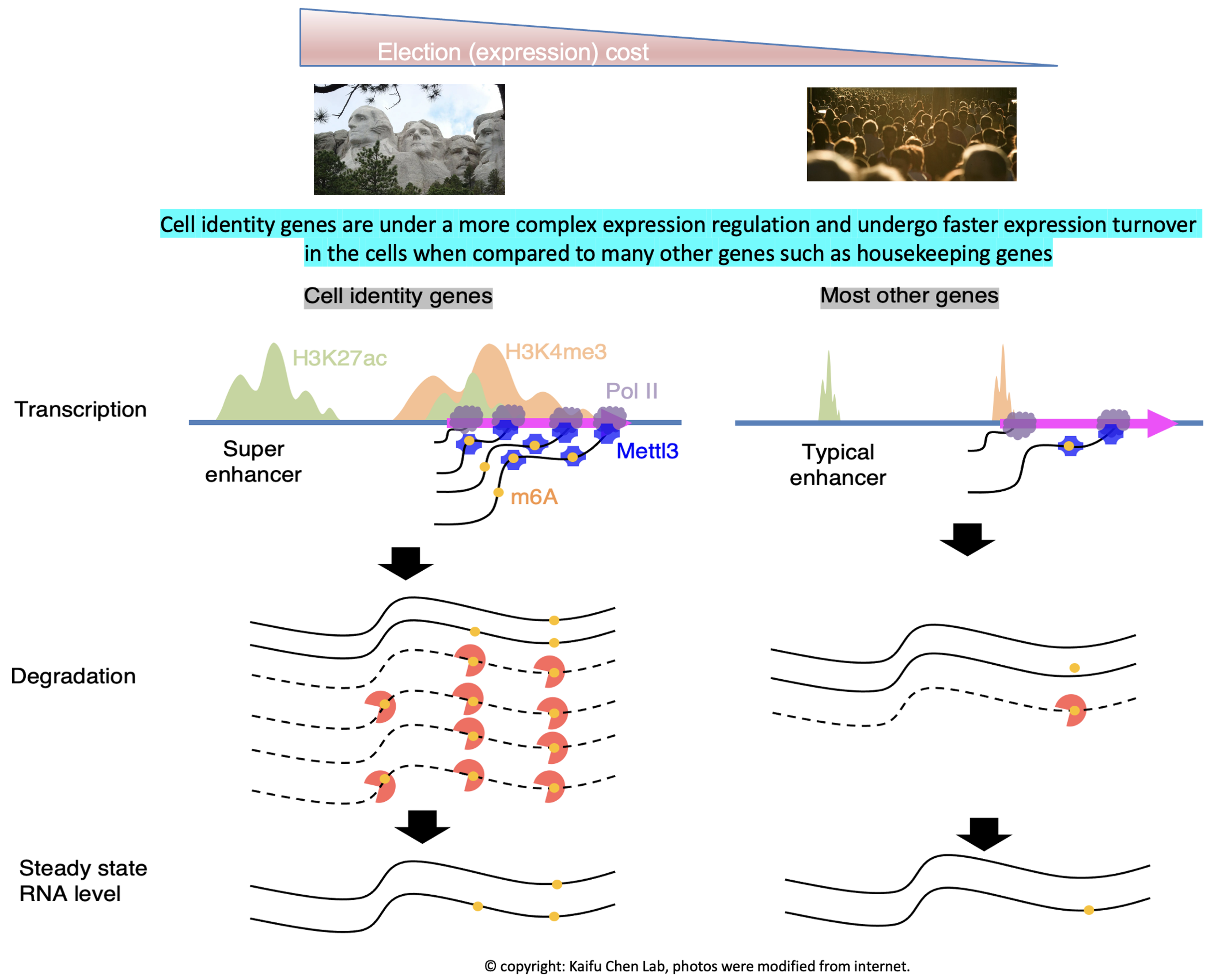
Our research findings offer a compelling rationale for targeting the intricate regulatory complexity governing cell identity genes in diseases linked to cell identity dysregulation. The heightened complexity of expression regulation at cell identity genes makes them particularly sensitive to manipulation of the associated molecular pathways. For instance, in our experiments, the knockout of the H3K4me3 methyltransferase MLL4 demonstrated that genes marked with broad H3K4me3 profiles were more susceptible compared to those with sharp H3K4me3 profiles. This vulnerability led to tumorigenesis and loss of normal cell identity (Molecular Cell, 2018). Additionally, the lower RNA stability observed for cell identity genes, attributed to their greater enrichment of co-transcriptional RNA m6A modification catalyzed by METTL3, revealed that knockdown of METTL3 had a more pronounced impact on RNA stability of cell identity genes compared to other genes (Nucleic Acids Research, 2023). Given that MLL4 and METTL3 may also influence many other expressed genes, there was a theoretical concern about potentially broad side effects when targeting these enzymes. However, our research suggests that a moderate dosage of MLL4 and METTL3 targeting may preferentially affect cell identity genes while exerting negligible effects on other genes. These findings provide a compelling justification for considering the therapeutic targeting of mechanisms govering the regulation complexity of cell identity genes in diseases associated with cell identity dysregulation.
